West China Hospital Sets a Benchmark with Huawei OceanStor Pacific as hWGS Results Can Be Analyzed Within Minutes
Two decades ago, the Human Genome Project (HGP) announced its initial results — the genome sequence and genetic map of the human body. Since then, advances in big data, storage, and computing technologies have ushered in a digital genomic era. Genome sequencing has advanced the study of genetics and has become an essential technique in the fight against viruses, especially severe acute respiratory syndrome (SARS) and more recently in the pandemic. Despite the huge advances in technologies and techniques, decoding the genome is much harder, especially without a specialist platform, preventing the spark that pushes our understanding of human life. Approx. 60,000 to 100,000 human genomes are distributed in 23 pairs of chromosomes in the nucleus and approximately three billion base pairs, making the scale to sequence and analyze at genome level coding incredibly difficult. Scientists spent US$3 billion and 13 years to complete that first human genome sequencing. Fast forward to 2021, and West China Hospital (WCH) of Sichuan University made a breakthrough in streamlining human whole genome sequencing (hWGS).
In September 2021, WCH, Huawei, and Sailegene jointly developed an acceleration analysis platform designed to handle the huge workloads of multiomics data. This platform shortens the 30x WGS for germline mutation analysis from 24 hours to just 7 minutes. It marks the world's first analysis of hWGS results to be completed in minutes, and changes the way in which enterprises analyze and decode multiomics data, signaling a new chapter in human life exploration.
WCH is a key medical and technological research base for national projects in China, ranking top in the Chinese Hospital Science and Technology Evaluation Metrics (STEM) for five consecutive years. STEM is published by Chinese Academy of Medical Sciences (CAMS). The hospital is now home to the West China Biomedical Big Data Center, an open platform that shares research and application of health and medical big data, and collects and analyzes biomedical data to improve clinical treatments. The center aims to build an efficient multiomics analysis platform that supports rapid large-scale analysis of next-generation sequencing in clinical practice.

Thanks to its many data sets, multiomics data analysis is the foundation of precision medicine and medical big data and provides a better understanding of the human system. Countries around the world are now investing in next-gen sequencing platforms for the big data of its populations. WCH, for example, started its WGS program in 2018 for approx. ten thousand elderly locals of different ethnic groups in western China and later in 2020 for 100,000 Chinese patients with rare diseases, using the high-performance genome analysis platform to undertake this project.
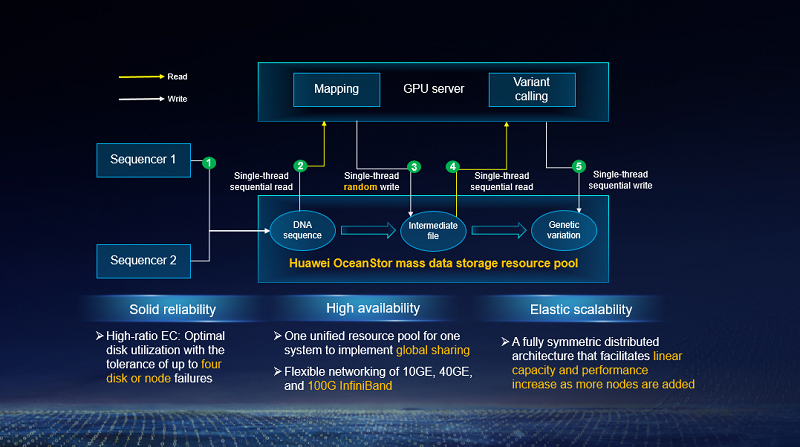
Genome sequencing aims to determine the complete sequence of genes from blood or saliva, and consists of three steps: extraction, analysis, and interpretation. It converts non-visualized bioinformation into library preparation (extraction) and reduces the deviation between text information and bioinformation by leveraging probability and statistics (analysis) for research (interpretation). Requiring a high-performance platform and sophisticated instruments, analysis involves file format conversion, decompression, gene splicing, sequence alignment, sequencing, deduplication, mutation detection, and joint genotype detection. Further, this study relies on the bioinformatic analysis system, making it a main focus for high-performance computing (HPC) solutions. WCH's challenges regarding genome sequencing were as follows:
First, a large amount of data needs to be processed. Typical genome sequencing projects generate TB-scale data. For example, an MGI's DNBSEQ-T7 sequencer produces 4.5 TB/24 h and 6 TB/30 h, and under full load can generate approx. 1.7 PB data annually. In addition, the intermediate files and results from bioinformatic analysis are about five times the raw data volume. The DNBSEQ-T7 requires 8.5 PB of effective storage capacity annually, making it a priority for cost-effective storage over extended periods while ensuring automatic management of online, offline, and archived data.
Second, the solution must be built for application-driven scientific computing workflows and the hybrid workloads of heterogeneous computing. During genetic data analysis, the varying nature of software and computing instances will produce I/O-, CPU-, and memory-intensive service needs, depending on their research objectives. The analysis and mining of mass genetic data require streaming processing and high-performance GPU and CPU heterogeneous computing clusters. In addition, sequence alignment, often performed during the analysis phase, requires a one-off import of mass data into the memory for processing, necessitating big memory capacity.
Third, superb storage performance is needed to facilitate the mass data transmission that typically imposes huge pressure on network bandwidth. Such transmissions cover share, read/write, and querying of huge data volumes, and demand high I/O bandwidth from both storage and other devices. The ideal solution must deliver a minimum throughput of 6 GB/s at a low latency to ensure data integrity.

This partnership makes use of the hospital's position as leading academic and industrial organization in multimodal omics analysis and application, Sailegene's experience in GPU-accelerated genome data analysis, and Huawei's leading position in developing high-performance storage and advanced data management systems. Together, the three parties produce an acceleration analysis platform for multiomics data. This platform empowers the biotechnology industry with mass data and innovative storage technologies, facilitating and even directing the digital transformation in the healthcare industry.
In this partnership, each party plays a specific role.
WCH provides high-performance software algorithms to analyze operation data and identify performance bottlenecks. It is responsible to design an optimization solution and a top-level architecture. The hospital also set up an R&D team to work with platform for multiomics data, as part of the globally leading high-performance genomic analysis platform.
Sailegene BaseNumber is an ultra-fast NGS data analysis platform that optimizes a single thread to multiple threads. It adds an intensive concurrent read/write mode for disks, to help improve both the write bandwidth (6 to 12 GB/s) and I/O throughput. BaseNumber also adds the fast cache synchronization to substantially accelerate the read and write of large files.
Huawei OceanStor Pacific mass data storage serves as the storage foundation that's to its strengths in performance. Because sequence alignment is a process in which big data sets are imported into the memory for processing, there is a huge demand for high single-thread storage bandwidth. Compared with WCH's legacy storage devices, OceanStor Pacific provides double the single-thread read bandwidth and four times the single-thread write bandwidth, at an aggregated bandwidth of 30 GB/s read and 25 GB/s write with just four nodes. Therefore, it significantly improves the performance of the multiomics joint innovation platform.

Through the comprehensive innovation of WCH, Sailegene, and Huawei in architecture, computing, and storage, the genome sequencing platform can complete analysis in just 7 minutes – a feat achieved for the first time globally. What is more, it completes the analysis at a rate 3.5 times faster than the legacy platform and 180 times faster than traditional solutions. At the 6th Biomedical Big Data and Intelligent Technology Application Summit, Dr. Yu Haopeng, data scientist at West China Biomedical Big Data Center, released the WHS-IMOAP high-performance genome analysis joint solution to the world, demonstrating the platform's capability to produce hWGS results within minutes. He announced that the age of multiomics big data has arrived. According to Dr. Yu, WCH introduced Huawei OceanStor Pacific, a converged analysis data foundation for diversified computing power, and Sailegene's GPU acceleration solution, into its acceleration analysis platform for multiomics data, helping shorten multiomics data analysis to minutes. He also pointed to this project as the defining moment in popularizing precision medicine in healthcare.
The seven-minute analysis for hWGS sets a benchmark for the global healthcare industry, and has far-reaching clinical and scientific research significance. In the future, the joint innovation platform will continue to process epigenomics, proteomics, and transcriptome data sets, and cover all practical clinical applications to help us better understand the human body.